Guided Growth of Nanowires Leads to Self-Integrated Circuits; The Pathway to Potato Poisons; Gene Decoding Obeys Road Traffic Rules
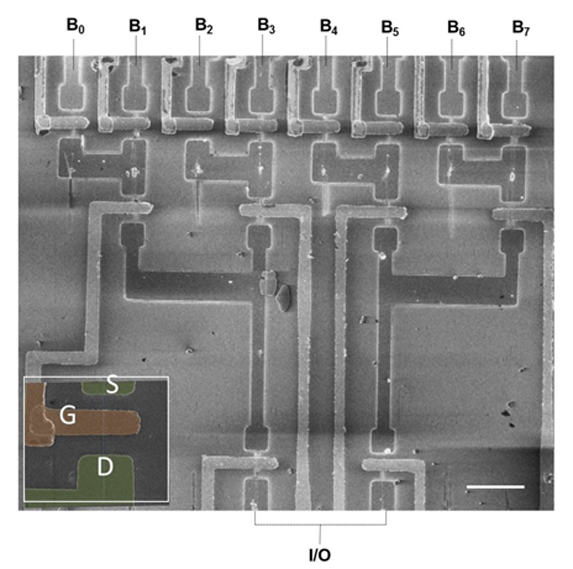
Scanning electron microscope (SEM) image of a logic circuit based on 14 nanowires. From the work of Prof. Joselevich.
Guided Growth of Nanowires Leads to Self-Integrated Circuits
Researchers working with tiny components in nanoelectronics face a challenge similar to that of parents of small children: teaching them to manage on their own. The nano-components are so small that arranging them with external tools is impossible. The only solution is to create conditions in which they can be “trusted” to assemble themselves.
Much effort has gone into facilitating the self-assembly of semiconductor nanowires, the basic building blocks of electronics, but until recently, success has been limited. Scientists had developed methods for growing nanowires vertically on a surface, but the resultant structures were short and disorganized. After growing, such nanowires need to be “harvested” and aligned horizontally; since such placement is random, scientists need to determine their location and only then integrate them into electric circuits.
A team led by Prof. Ernesto Joselevich of the Weizmann Institute’s Department of Materials and Interfaces has managed to overcome these limitations. For the first time, the scientists have created self-integrating nanowires whose position, length, and direction can be fully controlled.
The achievement, reported today in the Proceedings of the National Academy of Sciences (PNAS), USA, was based on a method developed by Prof. Joselevich two years ago for growing nanowires horizontally in an orderly manner. In the present study — conducted by Prof. Joselevich with Dr. Mark Schvartzman and David Tsivion of his lab, and Olga Raslin and Dr. Diana Mahalu of the Department of Physics of Condensed Matter — the scientists went further, creating self-integrated electronic circuits from the nanowires.
First, the scientists prepared a surface with tiny, atom-sized grooves and then added to the middle of the grooves catalyst particles that served as nuclei for the growth of nanowires. This setup defined the position, length, and direction of the nanowires. They then succeeded in creating a transistor from each nanowire on the surface, producing hundreds of such transistors simultaneously. The nanowires were also used to create a more complex electronic component — a functioning logic circuit called an Address Decoder, an essential constituent of computers. These ideas and findings have earned Prof. Joselevich a prestigious European Research Council Advanced Grant.
“Our method makes it possible, for the first time, to determine the arrangement of the nanowires in advance to suit the desired electronic circuit,” Prof. Joselevich explains. The ability to efficiently produce circuits from self-integrating semiconductors opens the door to a variety of technological applications, including the development of improved LED devices, lasers, and solar cells.
Prof. Ernesto Joselevich’s research is supported by the Carolito Stiftung and the European Research Council.
The Pathway to Potato Poisons
In 1924, Science magazine reported on a fatal case of potato poisoning: James B. Matheney of Vandalia, Illinois, had gathered about one and a half bushels of tubers, which turned green due to sunlight exposure. Two days after eating the potatoes, most of his family — wife, two daughters and four sons — showed symptoms of poisoning; the only exceptions were James himself, who didn’t eat the potatoes, and a breast-fed baby boy. His wife, aged 45, died a week later, followed by their 16-year-old daughter. The other five members of the family recovered.
Although such fatalities are rare among human beings, farm animals often get sick or die after eating green potatoes. Symptoms include damage to the digestive system as well as loss of sensation, hallucinations, and other neurological disturbances. Death can be caused by a disruption of the heartbeat. The culprits are the toxic substances solanine and chaconine; while they protect the tubers from insects and disease, their concentration rises sharply with exposure to light or during sprouting.
Solanine and chaconine belong to the large family of glycoalkaloids, which includes thousands of toxins found in small amounts in other edible plants, including tomatoes and eggplant. These substances have been known for over 200 years, but only recently has someone — Prof. Asaph Aharoni of the Department of Plant Sciences — begun to unravel how they are produced in plants. He and his team have mapped out the biochemical pathway responsible for manufacturing glycoalkaloids from cholesterol. Their findings will facilitate the breeding of toxin-free crops and the development of new crop varieties from wild strains that contain such large amounts of glycoalkaloids, they are currently considered inedible. On the other hand, causing plants to produce glycoalkaloids if they don’t do so naturally, or increasing their glycoalkaloid content, can help protect them against disease.
Two years ago, in research reported in The Plant Cell, the scientists identified the first gene in the chain of reactions that leads to the production of glycoalkaloids. Now, in a new study published recently in Science, they have managed to identify nine other genes in the chain by using the original gene as a marker and comparing gene expression patterns in different parts of tomatoes and potatoes. Disrupting the activity of one of these genes, they found, prevented the accumulation of glycoalkaloids in potatoes and tomatoes. The team then revealed the function of each of the genes and outlined the entire pathway, consisting of 10 stages, in which cholesterol molecules turn into glycoalkaloids.
An analysis of the findings produced an intriguing insight: Most of the genes involved are grouped on chromosome seven of the potato and tomato genome. Such grouping apparently prevents the plants from passing on to their offspring an incomplete glycoalkaloid pathway, which can result in the manufacture of chemicals harmful to the plants.
The research was conducted by postdoctoral fellow Dr. Maxim Itkin, who worked with Dr. Uwe Heinig, Dr. Oren Tzfadia, Pablo D. Cardenas, Dr. Samuel Bocobza, Dr. Sergey Malitsky, and Dr. Ilana Rogachev of Prof. Aharoni’s lab, as well as Dr. Tamar Unger of the Israel Structural Proteomics Center at the Weizmann Institute and scientists from the National Chemical Laboratory in Pune, India; the Hebrew University of Jerusalem; and the Wageningen University, the Netherlands.
Prof. Asaph Aharoni’s research is supported by the Clore Center for Biological Physics; the Kahn Family Research Center for Systems Biology of the Human Cell; the Tom and Sondra Rykoff Family Foundation; Roberto and Renata Ruhman, Brazil; the Adelis Foundation; the Leona M. and Harry B. Helmsley Charitable Trust; the Minna James Heineman Stiftung; and the Raymond Burton Plant Genome Research Fund. Prof. Aharoni is the incumbent of the Peter J. Cohn Professorial Chair.
Gene Decoding Obeys Road Traffic Rules
One of life’s most basic processes — transcription of the genetic code — resembles road traffic, including traffic jams, accidents and a police force that controls the flow of vehicles. This surprising finding, reported recently by Weizmann Institute researchers in Nature Communications, might facilitate the development of a new generation of drugs for a variety of disorders.
Transcription indeed involves a step resembling the motion of a vehicle: Enzymes “ride” along gene “tracks,” creating molecules that will later be translated into the various proteins involved in the life of the cell. In the new study, a research team headed by Prof. Rivka Dikstein of the Department of Biological Chemistry has found that, just as on the road, maintaining a reasonable distance between the vehicles — that is, the transcribing enzymes — is the surest way to reach a destination safely. In addition to Prof. Dikstein, the team included Dr. Nadav Marbach-Bar, Amitai Ben-Noon, Shaked Ashkenazi, Ana Tamarkin-Ben Harush, Dr. Tali Avnit-Sagi, and Prof. Michael Walker.
The scientists tracked the transcription of genes coding for tiny regulatory molecules called microRNAs. Working with human cells, they experimented with different rates of transcription: a high rate, in which the enzymes are launched in bursts, and a low one, in which the enzymes are launched individually, at greater intervals. The experiments yielded a paradoxical finding: When the transcription enzymes were launched in bursts, the amount of the resultant microRNA dropped; conversely, when the enzymes were launched at greater intervals, production of microRNA was more efficient.
It turned out that when the enzymes were launched in bursts, one rapidly following the next, they ended up in a traffic jam: When the first enzyme paused at a “road bump” — a molecular signal that creates a pause in transcription — the enzymes that followed crashed into it, falling off the gene. Naturally, such “traffic accidents” reduced the amount of resultant microRNA. In contrast, when the enzymes were launched one by one, they maintained a safe distance: Each had sufficient time to slow down at the “bump” and to succeed at creating a microRNA molecule. In other words, the lower rate of release of individual enzymes proved to be a more efficient method for creating microRNAs.
Because these findings shed new light on the manufacture of microRNAs, they might help in the design of drugs based on these molecules. Discovered relatively recently — the 1990s — microRNAs hold great promise for serving as future therapeutics because they can help control gene expression — for example, by blocking the activity of cancer-causing genes. This ability is particularly valuable when a molecular process needs to be manipulated at the deepest possible level, inside the cell nucleus.
In a more fundamental sense, the new study helps reveal how transcription is regulated. For example, the study has shown that in inflammation, when the body is threatened with invasion by a virus or bacterium, the release of anti-inflammatory microRNAs is temporarily suspended. The suspension occurs because inflammation increases the launch rate of transcription enzymes, creating traffic jams that reduce the production of the microRNA. This reduction, in turn, “buys time” for the inflammation, giving it a chance to perform its healing function before it is terminated by the microRNA.
Finally, this study helps explain an earlier finding in Prof. Dikstein’s lab: In longer genes, transcription enzymes tend to be launched at a low rate; that is, at great intervals. The longer the gene, the greater the risk that it has molecular “bumps” that can create traffic jams, derailing transcription. Therefore, transcription enzymes riding along such genes at a lower rate can do their job more efficiently than the enzymes launched in rapid bursts.
Prof. Rivka Dikstein’s research is supported by the Pearl Welinsky Merlo Foundation Scientific Progress Research Fund; the Yeda-Sela Center for Basic Research; the Wolfson Family Charitable Trust; and the Y. Leon Benoziyo Institute for Molecular Medicine. Prof. Dikstein is the incumbent of the Ruth and Leonard Simon Professorial Chair of Cancer Research.
Prof. Michael Walker’s research is supported by the Adelis Foundation and the Falconhead Charitable Foundation, Inc. Prof. Walker is the incumbent of the Marvin Myer and Jenny Cyker Professorial Chair of Diabetes Research.
