New Weizmann Institute technology speeds up DNA "rewriting" and measures the effects of the changes in living cells
REHOVOT, ISRAEL—May 31, 2012—Our ability to "read" DNA has made tremendous progress in the past few decades, but the ability to understand and alter the genetic code – that is, to "rewrite" the DNA-encoded instructions – has lagged behind. A new Weizmann Institute study advances our understanding of the genetic code: it proposes a way of effectively introducing numerous carefully planned DNA segments into genomes of living cells and of testing the effects of these changes. The study is being reported in the June issues of Nature Biotechnology and Nature Genetics.
Until now, changing the DNA sequence has been a slow and labor-intensive process. It took several weeks to alter just one DNA region at a time; testing the effects of each of these changes took even longer. In the new study, Weizmann Institute scientists have developed a technology that makes it possible to simultaneously introduce tens of thousands of DNA regions into tens of thousands of living cells – each region in a separate cell – in a planned and systematic manner, and to measure the results of each such change with great precision and within a single experiment.
"This fast method will significantly advance our ability to understand the ‘language' of DNA," says research team leader Prof. Eran Segal of the Weizmann Institute's Department of Computer Science and Applied Mathematics and Department of Molecular Cell Biology. "Reading out a person's entire genome is already a manageable task, but what exactly is written in that genome? After all, a genome looks like a lengthy string of letters whose meaning is for the most part obscure. Just deciphering the DNA letters is like trying to understand a foreign language by listening to it being spoken. Our method will help us identify DNA ‘words' and understand their meaning."
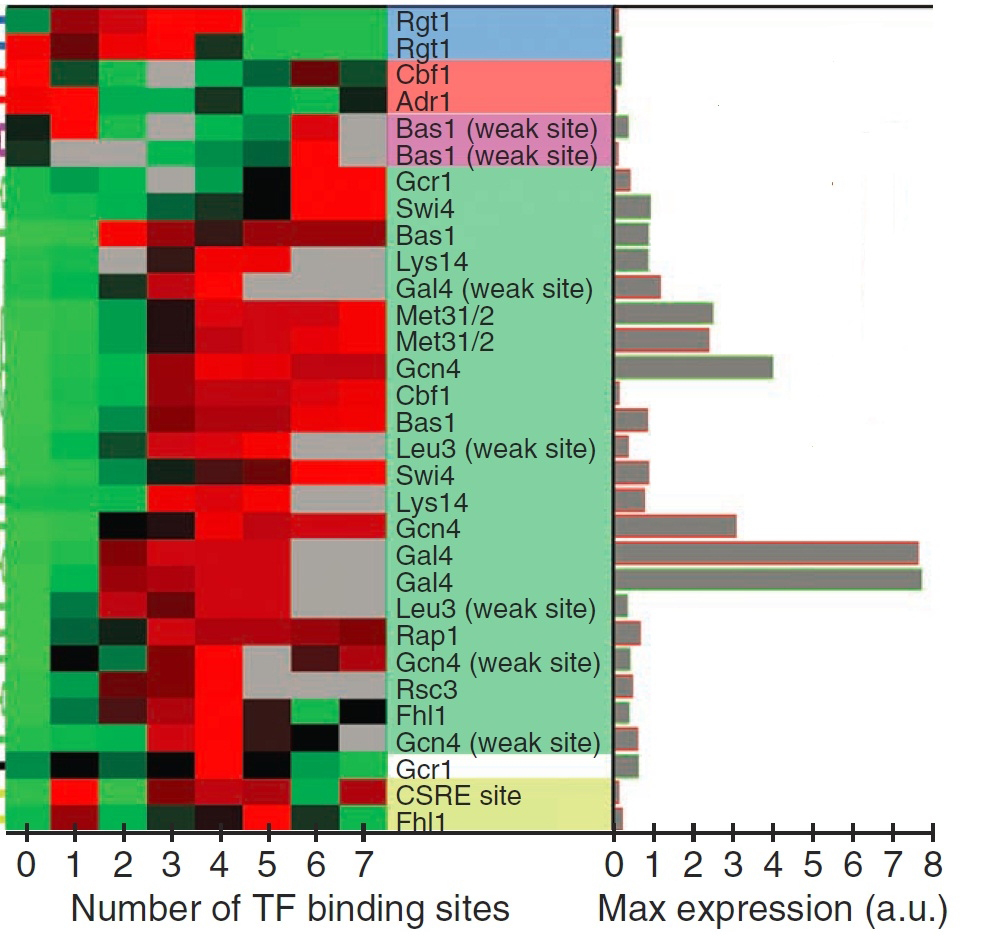
This graphic shows the relationship between gene activity and the number of binding sites for regulatory proteins in the gene's control region. The red-green scale shows gene activity levels, with red indicating high activity. The grey bars show the maximal level of gene activity achieved by each type of regulatory protein.
Understanding what's written in the DNA might help us interpret, among other things, how genotypic differences among people generate observable differences among them, from the way we look to the way our cells function. Thus, for example, it might be possible to clarify which genetic differences are responsible for the development of various diseases in certain individuals. The Weizmann Institute technology can also lead to improved genetic therapies based on introducing new genes or improved regulatory sequences into cells in order to repair genetic defects.
In the present study, the scientists investigated a vital aspect of the DNA language: how the control of gene expression is encoded in the DNA – that is, the instructions determining the level of activity of each gene in the genetic code. Since gene activity levels have crucial effects on cell function, this question, considered one of the central in molecular biology, has been studied for decades. The new technology has enabled the scientists to isolate and test the effects of various parameters on gene activity levels: for example, how a gene's activity level is affected by the gene's distance from its regulatory sequence. The researchers have managed to elucidate how various parameters define the regulatory "language" and to demonstrate how deliberate changes in the genetic sequence affect these parameters in a way that alters the level of a gene's activity in a predictable manner.
The new method consists of four steps that combine existing technologies in an innovative manner. The steps are: creation of 50,000 different genetic sequences on DNA chips; massive insertion of these sequences into cells at the same time; sorting the cells with the help of a sorting machine that senses the expression levels of a "reporter" gene; and high-throughput parallel DNA sequencing.
Taking part in the study were Weizmann Institute graduate students Eilon Sharon, Tali Raveh-Sadka, and Michal Levo; research assistant Dr. Yael Kalma and research associate Dr. Adina Weinberger; as well as Dr. Zohar Yakhini from the Technion – Israel Institute of Technology and Agilent Laboratories, Santa Clara, California.
Prof. Eran Segal's research is supported by the Cecil and Hilda Lewis Charitable Trust; the Carolito Stiftung; the Kahn Family Research Center for Systems Biology of the Human Cell; and the European Research Council.
